What health professions in oncology needs to know about pharmacogenomics?
WCRJ 2014; 1 (1) : e90
Topic: Pharmacogenomics
Category: Review
Abstract
BACKGROUND: Pharmacogenomic offers the promise to ranking an oncologic disease into genetic sub-categories, allowing bespoke tailoring of medicine to maximize therapeutic effects and to reduce adverse drug response. This new feature requires for health professionals to have competencies not only for their discipline, but also for the skills on why, when, and how that pharmacogenomic knowledge should be applied to improve personalized therapies for their patients. Current opinion on basic competences of health professions includes knowledge and skills on two fundamental features: a) genetics of disease, to allow the understanding and the identification of diseases associated to genetic variations, and to facilitate the application of new genomic tests; and b) ethical, social and economical implications that are fundamental to identify those factors that might contribute to a successful integration of pharmacogenomics into international health and public policy.
AIM: Briefly, we described i) current knowledge on genetic variations that interact with therapies, the need to detect them with the most common available methods; and ii) ethical, social and economic issues related to pharmacogenomic testing and recording of genetic information (e.g., critical evaluation of the development of new tests, privacy, the current absence of public reimbursement, etc).
CONCLUSIONS: These issues should be useful recommendations for academic institutions and educational programs to prepare health professionals in pharmacogenomic field with the necessary abilities for their future practice.
INTRODUCTION
The developments in pharmacogenetics and pharmacogenomics (PGx), detailed information about human genome made available and the genetic basis for success/failure of pharmacotherapy in oncology have being studied 1.
Pharmacogenetic knowledge is rapidly developing and changing; it is imperative that healthcare professionals keep abreast of advances and clinical indications. The current knowledge of health professions regarding PGx is still low. There exists an acute lack of education of both physicians and pharmacists regarding pharmacogenetics and personalized care 2. Academic curricula are slowly including teaching of this field in their courses. Healthy institutions and academic organizations must play a central role in educating health professionals on the best use for applications of advancing pharmacogenomics research applied to oncology, and in articulating on the role of physicians and pharmacists in the development and use of gene-based therapies, as well as in making treatment choices as the result of available patient-specific genetic information 3.
The large number of drug options also means that physicians are often spoilt for choice, and have a low threshold to consider alternative therapies when toxicity becomes unmanageable.
However, it is often forgotten that genetic testing is not only predictive for treatment related toxicity or allows for dose adjustment, but also determines response or lack thereof. It is frequently imperative that must be done before treatment, as giving inappropriate treatment may result in an outcome poorer than the alternative 4. A ‘treat-and-see’ approach has ethical and legal implications in this era where genetic testing is readily available. It delays and even potentially deprives patients of appropriate treatment, and deterioration is often rapid without it. Moreover, we think that genetic testing could have a key role for the treatment choice in the so called frail patients (i.e. elderly and HIV-positive patients) for whom the efficacy and especially the toxicity profile are important aspects 5 , 6.
However, one should keep in consideration that it will not be feasible to conduct randomized trials on each and every diagnostic test, and the economic value of such tests can be modelled using decision analysis techniques.
The goal of this review is to provide information (in terms of knowledge-base in genetics, ethical, social and economic) for the health profession about the genetic variations implicate in oncologic pharmacotherapy.
GENETICS COMPETENCIES
Pharmacogenomic approaches have been applied to many existing therapeutic agents in an effort to identify relevant inherited variations that may better predict patients’ response to treatment. Genetic variations, which can alter the protein expressions and/or amino acid sequence of the encoded proteins, include nucleotide repeats, insertions, deletions, translocations and Single Nucleotide Polymorphisms (SNPs).
Such genetic polymorphisms in drug metabolizing enzymes like the Cytochrome P450 family 7; transporters like Multidrugs Receptors-1 8; and molecular targets, have been actively explored with regard to functional changes in phenotype (altered expression levels and/or activity of the encoded proteins) and their contribution to variable drug response. The following Table 1 describes some clinically relevant examples of genetic defects illustrating the relevance of PGx in optimizing pharmacotherapy, as a way to enhance efficacy and safety.
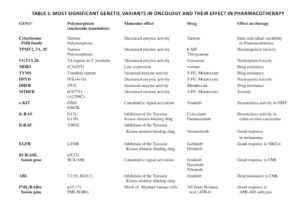
Abbreviations: TPMT = thiopurine methyltransferase; UGT1A1 = UDP-glucuronosyltransferase 1A1; MDR1 = multidrug resistance 1; TYMS = thymidylate synthase; DPYD= Dihydropyrimidine Deaminase; DHFR= Dihydrofolate reductase; MTHFR = 5,10-methylene tetra hydrofolate reductase; EGFR= Epidermal Grow Factor Receptor; 5-FU = 5-fluorouracil; 6-MP = 6-mercaptopurine; AML= Acute Myeloid Leukemia; NSCLC= Non-Small Cell Lung Cancer; CML= Chronic Myeloid Leukemia;
The present list is not meant to be whole comprensive.
a Genes are available for genotyping test or under consideration for clinical diagnostics
For example the new generation of anticancer drugs have high specificity toward tumour cells, provide a broader therapeutic window with less/low toxicity in comparison with conventional chemotherapies; therefore, these drugs represent a new and promising approach to targeted cancer therapy. These new drugs are designed to interfere with a specific molecular target, usually a protein with a critical role in tumour growth or progression (i.e. tyrosine kinase). There are multiple types of targeted therapies available, including monoclonal antibodies, antisense inhibitors, and inhibitors of tyrosine kinase. Obviously, many of these new drugs set up a selective pressure for tumour cells that can survive and proliferate in its presence. The same basic principle seems to be true for protein kinase inhibitors. The best understanding of this problem at a molecular level comes from studies on imatinib resistance in Chronic Myelogeneous Leukaemia (CML) patients carrying BCR/ABL fusion gene. These imatinib-resistant clones, consisting a single nucleotide mutation in ABL Kinase domain (with consequent amino acid substitution), are successfully suppressed by second-generation Tyrosine kinase inhibitors (i.e. Dasatinib, Nilotinib), still active on almost all imatinib-resistant mutants 9.
Similarly to imatinib, other two biological drugs (Gefitinib and Erlotinib) showed clinical activity in a subset of patients affected by Non Small Cell Lung Cancer (NSCLC). The mechanism of action for both drugs is the selective inhibitions of the kinase activity of epidermal growth factor receptor (EGFR) 10. Recently, it has been reported in NSCLC patients that specific point mutation of EGFR gene in tumour cells select Gefitinib-responders’ patients (EGFR mutated), from non-responders (EGFR wild type) 11. The availability of this kind of biomarkers is currently useful tool for predicting resistance to specific drug therapy.
GENOTYPING METHODS AND ECONOMICS COMPETENCIES
The technology platform needed for genotyping is different; it does depend from type of mutation, acquired genetic change, or the analysis of inherited SNPs. No single genotyping platform stands out as ideal and rational selection of the best methods to detect them is dependent from the specifics aims of different laboratories 12. Furthermore, the most popular technologies currently used in specialized laboratories, focus on the transition from research setting to clinical laboratory as previously discussed by other authors 13.
As genomics-based technologies are widely introduced in clinical laboratories testing setting, the risks of mishandling or misinterpreting data from patient’s sample analyses becomes a significant consideration with especially dramatic consequences where the test becomes commercially available to the public 14.
Generally, genotyping is performed either by custom clinical laboratories or academic referenced laboratories, as well as by using commercial kits (when available). In the USA, diagnostics products are regulated by the Food and Drug Administration (FDA), whereas diagnostic services are under the rules of the Clinical Laboratory Improvement Act (CLIA). In Europe this field is covered by in vitro Diagnostic (IVD) directive, without a distinction between commercial products (used by laboratories) and diagnostics service. In both circumstance, a voluntary list of international laboratories (with CLIA certification in the US and CE mark in Europe) that perform genetic tests can be found on the National Institute of Health-funded website named GeneTests™ [http://www.ncbi.nlm.nih.gov/sites/GeneTests/lab?db=GeneTests], although only a small minority of genetic tests listed on this site are PGx tests. Clinical laboratories may develop and validate tests in-house (“home-brew”) and perform them as a laboratory service; which may further reduce the cost of analysis 15 , 16. Furthermore, laboratory-developed molecular tests are in contrast with patented kit manufactured by company biotech. The impact of these commercial controversial was low studied by Academic Institution, except for the positive model of the US Department of Health and Human Services (HSS) [http://oba.od.nih.gov/SACGHS/]. In the field of oncology, there are new 38 genomic tests approved by FDA since 2006.

Appendix to table 2
Issues listed in table 2, derives in part, from, National Coalition for Health Professional Education in Genetics (//wikigenetics.org/index.php/NCHPEG-Principles_of_Genetics_for_Health_Professionals) and in part from, databases available to the genetic community with a wide range of aims and scopes including: i) those presenting guidelines on pharmacogenomics related to government policy such as Food and drug Administration (www.fda.gov/cder/genomics/default.htm) and European Medicine Agency (www.emea.europa.eu/pdfs/human/ich/43798606en.pdf); ii) those provide a genetic row data such as Genbank (www.ncbi.nlm.nih.gov/Genbank/index.html); and iii) those providing higher level structure and annotation such as Pfam (www.sanger.ac.uk/Software/Pfam). Other types of large-scale data resource used in Pharmacogenetic testing include publication databases such as Pharmacogenomics Knowledge Base (www.pharmgkb.org/index.jsp) and disease/gene information resources and tools such as OMIM (www.ncbi.nlm.nih.gov/sites/entrez?db=omim/) or Orphanet. (www.orpha.net). The table lists of FDA-approved drugs with pharmacogenomic information in their labels are available at www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm. Clearly there are overlaps between many of these database and an attempt at categorize them to any detailed degree may be difficult and unproductive.
Website accessing: until February 2014
ETHICAL, SOCIAL AND COST-EFFECTIVENESS
Pharmaceutical and Biotech companies frequently develop their own clinical pharmacokinetic and pharmacodynamic tests for new drug studies. They are required to have validated assays for human clinical phase III trials complying with current Good Clinical Practice guidelines for FDA or EMEA submission purposes. This involves testing patients as a potential recipients before the administration of the drug. This difference poses an ethical dilemma for pharmaceutical companies, especially if inadequate testing excludes some patients who might benefit from receiving the drug or, conversely, long-term dosing continues with a treatment that does not have good clinical efficacy 17. Pharmaceutical companies should be involved with the initial development of PGx assays because they have the primary data and information necessary for this stage of assay development. However, this assay development activity should be transferred to outside referenced laboratories, clinical core laboratories in academic health centers, or established Clinical Research Organizations when research and development transit into clinical application because these independent external sites are able to handle this function 18.
Drug selection based on genetic assessment may be considered confidential information. Currently, PGx testing may provide detailed genetic information necessary for health professions to prescribe the correct drug and its dose. The skills of health operator must be orientated to maintain the confidentiality and security of patient’s health records. In this field, the clinical laboratories could be the most proficient means to protect patient/physician confidentiality.
Reimbursement or payment for genetic testing is another topic of considerable consequence that is already creating controversy among health maintenance organizations, healthcare providers, and the patients themselves. One can predict, however, that health insurance companies will be very interested in patient PGx testing to document the proper dosing of expensive prescription drugs and hence reduce the incidence or risks of adverse drug reactions. It will be interesting to see whether insurers will consider PGx testing to be a cost-effective alternative to the current trial-and-error approach to dosage regulation. However, if the detection of these genetic variants is routinely incorporated either into clinical practice or large clinical trials, knowledge concerning the predictive value of PGx which will eventually enable the individualization of optimized therapy could be gained 19. However, we still need a precise demonstration that PGx tests offer an added value, in terms of relative cost and benefit.
Furthermore, trials evaluating the pharmacoeconomic impact of genotyping testing before therapy will likely provideanswers for policy making in the merging of PGx testinginto clinical practice. The primary aim of a cost–effectiveness analysis is to provide sufficiently robust information for decision-makers to allocate resources to healthcare interventions. Overviews of cost-effectiveness studies on PGx technologies are now available 20. A relevant example is the National Institute for Health and Clinical Excellence (NICE). NICE forms a Diagnostic Advisory committee, which is willing to stimulate Pharma and Academic communities to produce a robust set of data, including design and data source in economic models of healthcare 21. Only few studies have addressed the cost-effectiveness of pharmacogenomics testing implication in clinical practice 22 included thiopurine S-methyltransferase (TPMT) genotyping prior to 6-mercaptopurine treatment in paediatric Acute Lymphoblastic Leukaemia (ALL); the mean calculated cost from 4 European countries was € 2100,00 per life-year considering low myelosuppression-related hospitalization; the cost for genotyping of TMPT mutation averaged around €150,00. Early outline of genotyping cost for “home brew” pharmacogenomic tests averaged about €20,00 per SNP 23.
CONCLUSION AND FUTURE OUTLOOK
The potential is enormous for pharmacogenomics to yield a powerful set of molecular diagnostics that will become routine tools by which pharmacists and physicians select the proper medications and doses for each individual patient. Instead of starting patients on the “average dose” that was found to be safe and effective in most patients in large clinical trials, pharmacogenomics has the potential to provide patient-specific data upon which the selection of drugs and doses can be individualized and optimized. Using the amount of DNA that can be isolated from just few milliliters of blood, it is possible to determine thousands of genotypes in diverse genes. So, taken together, the process will be to collect a single blood sample from each patient, submit an aliquot of the sample to a reference laboratory for analysis of a panel of genotypes, and test for those established to be important determinants of drug disposition and effects. The results of this specific panel of genetic variants would be electronically deposited into a secured database, into and out of which data can be accessed only with the patient’s authorization (to her/his health care professionals). The results of these tests will not be simply a list of gene SNPs, but rather a report formatted and interpreted according to the patient’s diagnosis and treatment options. For example, the report could be a recommended algorithm for the selection of antineoplastic, starting with those most likely to be effective and well tolerated, based on the patient’s genotypes for the panel of genes known to be significant determinants of the disposition and effects of chemotherapeutic medications (i.e. DPYD mutation and 5-Fluorouracil administration) 24. As patients experience additional illnesses, additional genotypes will be characterized and the data added to the same secured database, to which the patient’s future physicians and pharmacists would be granted access as needed to make treatment decisions. Of course, pharmacogenomic will not replace the more conventional biochemical tests that are now routinely used to assess organ function and disease progression; rather they will complement these contemporary tests and provide additional tools for selecting the optimal medications for individual patient’s treatments. Furthermore, genotyping will not obviate the need for follow up assessment of response, adherence to treatment, or drug interactions, which will continue to be important clinical responsibilities of health professionals.
Promise in the next few years, the needs to detect new genomic alterations in therapy will drive diagnostics companies to develop new tests able to produce results for tailoring patient’s treatment.
Conclusions
With the increasing number of novelgenetic markers being identified and validated, pharmacogenomics will make the practice of health profession and medicine should be less an art and more a science.
Aknowledgements: the authors are grateful to Dr. O. Barletta from the “Italian Association of Pharmacogenomics and Molecular Diagnostics” for the invaluable bibliography research and Mrs. Paola Favetta from “GORI” for her expert assistant in the preparation and correction of the manuscript.
Conflict of interest statement
Authors’ conflict of interest disclosure: The authors stated that there are no conflicts of interest regarding the publication of this article.
Research funding: None declared.
References
- McCarthy JJ, McLeod HL, Ginsburg GS. Genomic medicine: a decade of successes, challenges, and opportunities. Sci Transl Med 2013; 5: 189. (back)
- McCullough KB, Formea CM, Berg KD, Burzynski JA, Cunningham JL, Ou NN, Rudis MI, Stollings JL, Nicholson WT. Assessment of the pharmacogenomics educational needs of pharmacists. Am J Pharm Educ 2011; 75: 51. (back)
- Di Francia R, Valente D, Catapano O, Rupolo M, Tirelli U, Berretta M. Knowledge and skills needs for health profession about pharmacogenomics testing field. Eur Rev Med Pharmaco Sci 2012; 16: 781-788. (back)
- McCarthy JJ, McLeod HL, Ginsburg GS. Genomic medicine: a decade of successes, challenges, and opportunities. Sci Transl Med 2013; 5: 189. (back)
- Berretta M, Cappellani A, Fiorica F, Nasti G, Frustaci S, Fisichella R, Bearz A, Talamini R, Lleshi A, Tambaro R, Cocciolo A, Ristagno M, Bolognese A, Basile F, Meneguzzo N, Berretta S, Tirelli U. FOLFOX4 in the treatment of metastatic colorectal cancer in elderly patients: a prospective study. Arch Gerontol Geriatr 2011; 52: 89-93. (back)
- Berretta M, Di Benedetto F, Bearz A, Simonelli C, Martellotta F, Del Ben C, Berretta S, Spina M, Tirelli U. FOLFOX-4 regimen with concomitant highly active antiretroviral therapy in metastatic colorectal cancer HIV-infected patients: a report of five cases and review of the literature. Cancer Invest 2008; 26: 610-614. (back)
- Aquilante CL, Lobmeyer MT, Langaee TY, Johnson JA. Comparison of cytochrome P450 2C9 genotyping methods and implications for the clinical laboratory. Pharmacotherapy 2004; 24: 720-726. (back)
- Panczyk M, Balcerczak E, Piaskowski S, Jamroziak K, Pasz-Walczak G, Mirowski M. ABCB1 gene polymorphisms and haplotype analysis in colorectal cancer. Int J Colorectal Dis 2009; 24: 895-905. (back)
- Bui MR, Hodson V, King T, Leopold D, Dai S, Fiolkoski V, Oakes S, Duke R, Apelian D, Franzusoff A, DeGregori J. Mutation-specific control of BCR-ABL T315I positive leukemia with a recombinant yeast-based therapeutic vaccine in a murine model. Vaccine 2010; 28: 6028-6035. (back)
- Gazdar AF. Tyrosine kinase inhibitors and epidermal growth factor receptor (EGFR) mutation in non-small cell lung cancer: to test or not to test? Medicine (Baltimore) 2010; 90: 168-170. (back)
- de Mello RA, Marques DS, Medeiros R, Araújo AM. Epidermal growth factor receptor and K-Ras in non-small cell lung cancer-molecular pathways involved and targeted therapies. World J Clin Oncol 2011; 2: 367-376. (back)
- Di Francia R, Frigeri F, Berretta M, Cecchin E, Orlando C, Pinto A, Pinzani P. Decision criteria for rational selection of homogeneous genotyping platforms for pharmacogenomics testing in clinical diagnostics. Clin Chem Lab Med 2010; 48: 447-459. (back)
- Aquilante CL, Zineh I, Beitelshees AL, Langaee TY. Common laboratory methods in pharmacogenomics studies. Am J Health Syst Pharm 2006; 63: 2101-2110. (back)
- Deverka PA. Pharmacogenomics, evidence, and the role of payers. Public Health Genomics 2009; 12: 149-157. (back)
- Di Francia R, Frigeri F, Berretta M, Cecchin E, Orlando C, Pinto A, Pinzani P. Decision criteria for rational selection of homogeneous genotyping platforms for pharmacogenomics testing in clinical diagnostics. Clin Chem Lab Med 2010; 48: 447-459. (back)
- Di Francia R, Berretta M, Catapano O, Canzoniero LM, Formisano L. Molecular diagnostics for pharmacogenomic testing of fluoropyrimidine based-therapy: costs, methods and applications. Clin Chem Lab Med 2011; 49: 1105-1111. (back)
- Payne K, Shabaruddin FH. Cost-effectiveness analysis in pharmacogenomics. Pharmacogenomics 2010; 11: 643-646. (back)
- Kirk R, Edwards LB, Aurora P, Taylor DO, Christie J, Dobbels F, Kucheryavaya AY, Rahmel AO, Hertz MI. Registry of the International Society for Heart and Lung Transplantation: eleventh official pediatric heart transplantation report–2008. J Heart Lung Transplant 2008; 27: 970-977. (back)
- Payne K, Shabaruddin FH. Cost-effectiveness analysis in pharmacogenomics. Pharmacogenomics 2010; 11: 643-646. (back)
- Payne K, Shabaruddin FH. Cost-effectiveness analysis in pharmacogenomics. Pharmacogenomics 2010; 11: 643-646. (back)
- Dhalla IA, Garner S, Chalkidou K, Littlejohns P. Perspectives on the National Institute for Health and Clinical Excellence’s recommendations to use health technologies only in research. Int J Technol Assess Health Care 2009; 25: 272-280. (back)
- Payne K, Shabaruddin FH. Cost-effectiveness analysis in pharmacogenomics. Pharmacogenomics 2010; 11: 643-646.). For example, van den Akker et al ((Van den Akker-van Marle ME, Gurwitz D, Detmar SB, Enzing CM, Hopkins MM, Gutierrez de Mesa E, Ibarreta D. Cost-effectiveness of pharmacogenomics in clinical practice: a case study of thiopurine methyltransferase genotyping in acute lymphoblastic leukemia in Europe. Pharmacogenomics 2006; 7: 783-792. (back)
- Di Francia R, Berretta M, Catapano O, Canzoniero LM, Formisano L. Molecular diagnostics for pharmacogenomic testing of fluoropyrimidine based-therapy: costs, methods and applications. Clin Chem Lab Med 2011; 49: 1105-1111.14 (back)
- Sween JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, Rongen GA, van Schaik RH, Schalekamp T, Touw DJ, van der Weide J, Wilffert B, Deneer VH, Guchelaar HJ. Pharmacogenetics: from bench to byte–un update of guidelines. Nature 2011; 85: 662-673. (back)
To cite this article
What health professions in oncology needs to know about pharmacogenomics?
WCRJ 2014; 1 (1) : e90
Publication History
Published online: 21 Mar 2014

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.